READ:
Robust Elicitation Algorithms for Discovering DNA Motifs
Using Fuzzy Self-Organizing Maps
Dianhui Wang and Sarwar Tapan
Introduction Being a hard c-means type algorithm, the learning algorithm of the classical SOM treats the data samples lying around the cluster boundaries by forcing them to belong to one of the clusters by its winning node selection process. This crisp-clustering approach can have a considerable negative impact on the DNA motif mining. Since, in crisp partitioning it is usually unavoidable to have some true but weak binding sites distributed among the neighbouring clusters of a true motif, especially in weak motif mining. READ framework addresses this problem by using a fuzzy-SOM approach that performs fuzzy c-means (FCM) type soft partitioning of the data space besides preserving the map topology. Hence, each motif-like cluster in a fuzzy-SOM map is able to consider the global associations of all the data samples in order to maximize the chances of attaching weak binding sites.
|
|
Download READ has been developed on a MATLB environment, and the executables are released for public use and can be downloaded from the link: download READ_release_version_1.rar The package contains the following files and folders:
|
|
Installation and usage: READ is expected to run as a standalone application on a computer that has a licensed MATLB environment installed on a Windows XP, Vista, or Windows 7 platform. If the computer does not have any MATLAB environment installed then the MATLAB Compiller Runtime (MCR) needs to be installed first. The MCR is provided with the READ package and can be installed by double-clicking on the MCRInstaller.exe file. It is however recommended for a computer with MATLAB installed that, the MCR should also be installed for version compatibility. |
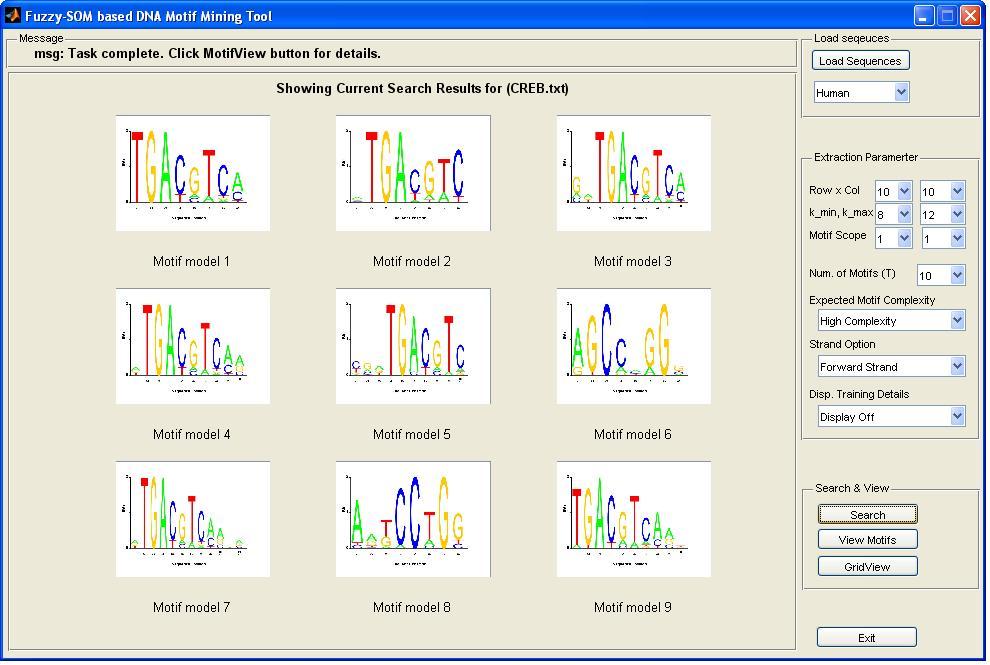
Sample Run: A sample motif mining result produced by READ on a real DNA dataset, e.g., CREB protein, can be as follows:
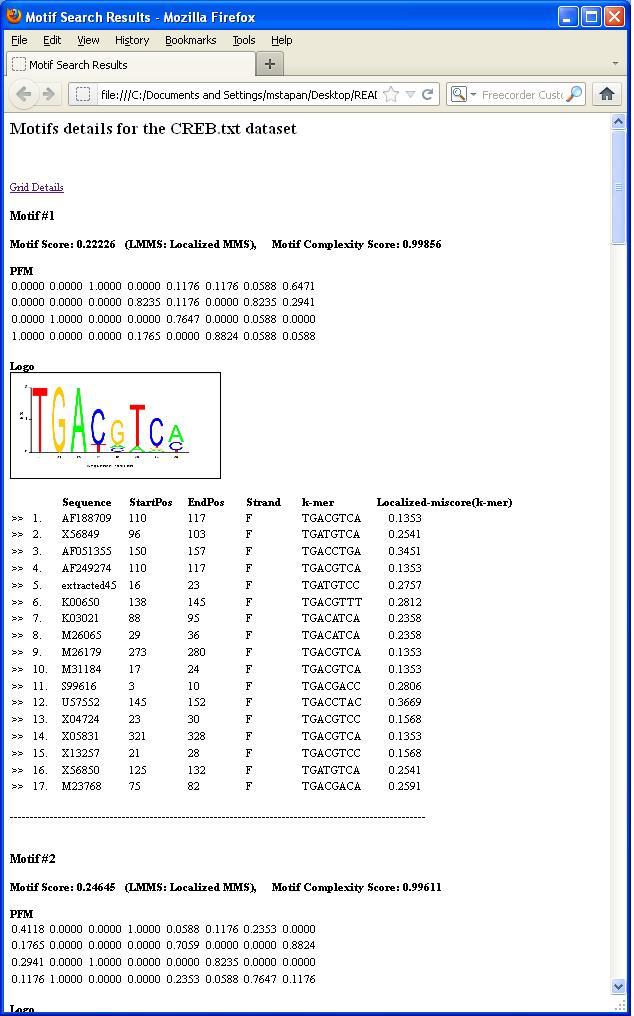
Then motif details can be viewed by clicking the "View Motif" button, which will generate a html page that might look like the following:
|